The cloning scheme that we will use to get single stranded oligos with no PCR handle overhangs from the oligo pool comes from this paper on 'MO-MAGE'. Note that there is detailed information in the supplement.
Their strategy was to amplify subpools of oligos as usual, but there are a few clever modifications. First, when amplifying, the reverse primer is modified to include a 5' phosphate group. This 5' end, i.e. the '-' strand will be selectively degraded by the lambda exonuclease (neb link). Also during that PCR it seems that the fwd primer has multiple 5' end PO bonds that selectively protect the '+' strand.
To cleave off the PCR handles, they used a uracil in the fwd primer to introduce a site for the USER enzyme, and they included a DpnII site in the reverse primer sequence. By annealing just the reverse primer you can create a double stranded template for DpnII, which will cleanly cleave off the 5' end of the '+' strand. See the diagram below:
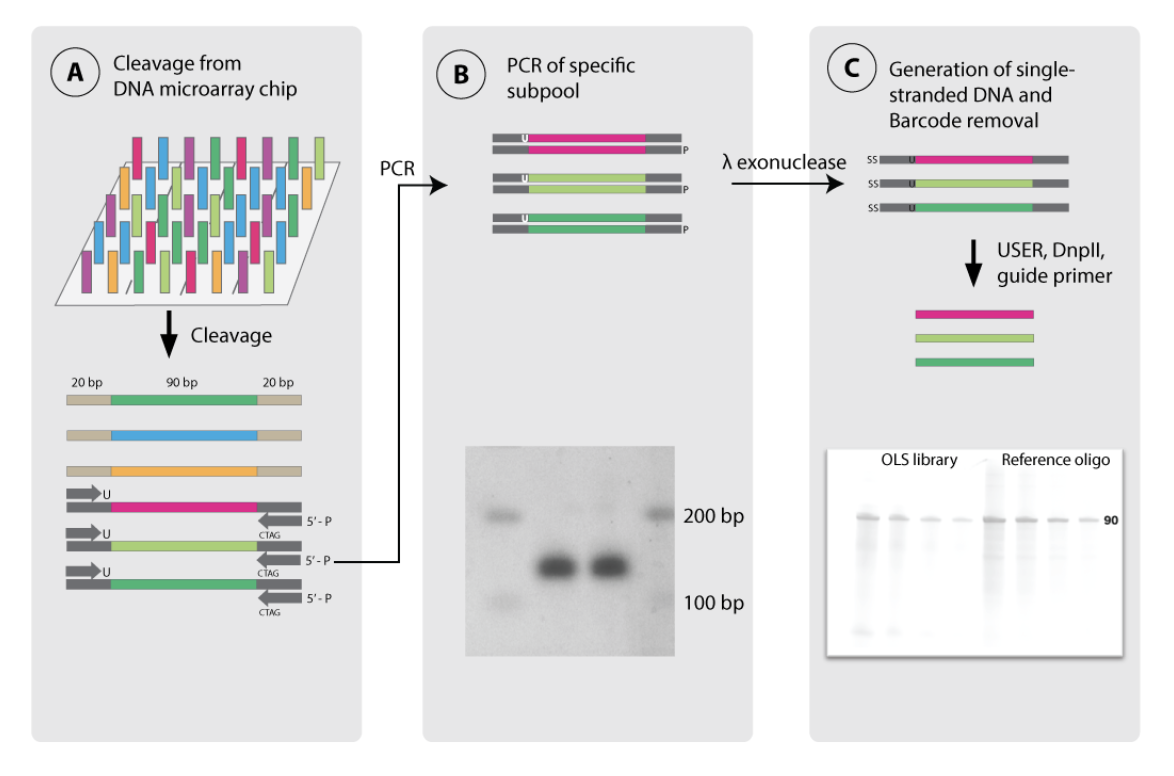
In this way, they got ssDNA oligos with no overhangs.
I will modify their approach slightly, mainly because the USER enzyme is quite expensive, and I would like to avoid it if possible. For my cloning scheme I will introduce two restriction sites: btsI-V2 and dpnII / mboI. DpnII seemed to work well enough in their scheme for cleaving the 3'end, but it requires a special buffer, so I will actually use MboI to start, which cleaves at the exact same sequence. Throughout this document I may refer to the dpnII site, but remember that downstream I will actually use MboI. I chose BtsI-V2 for the 5' end because it does not leave a 3' overhang I can still include a T as the 5' overhang, allowing us to use the USER enzyme if this approach fails.
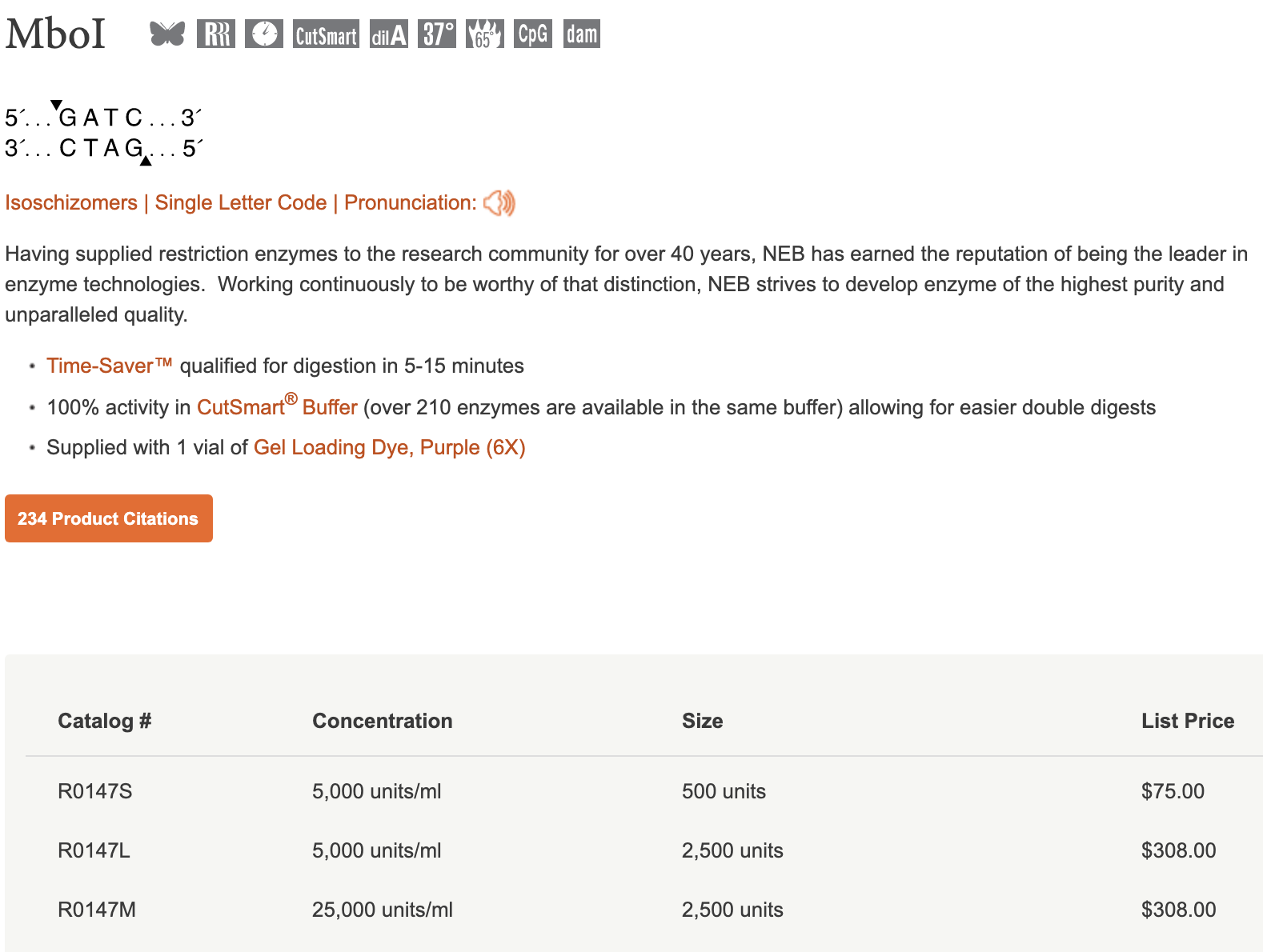
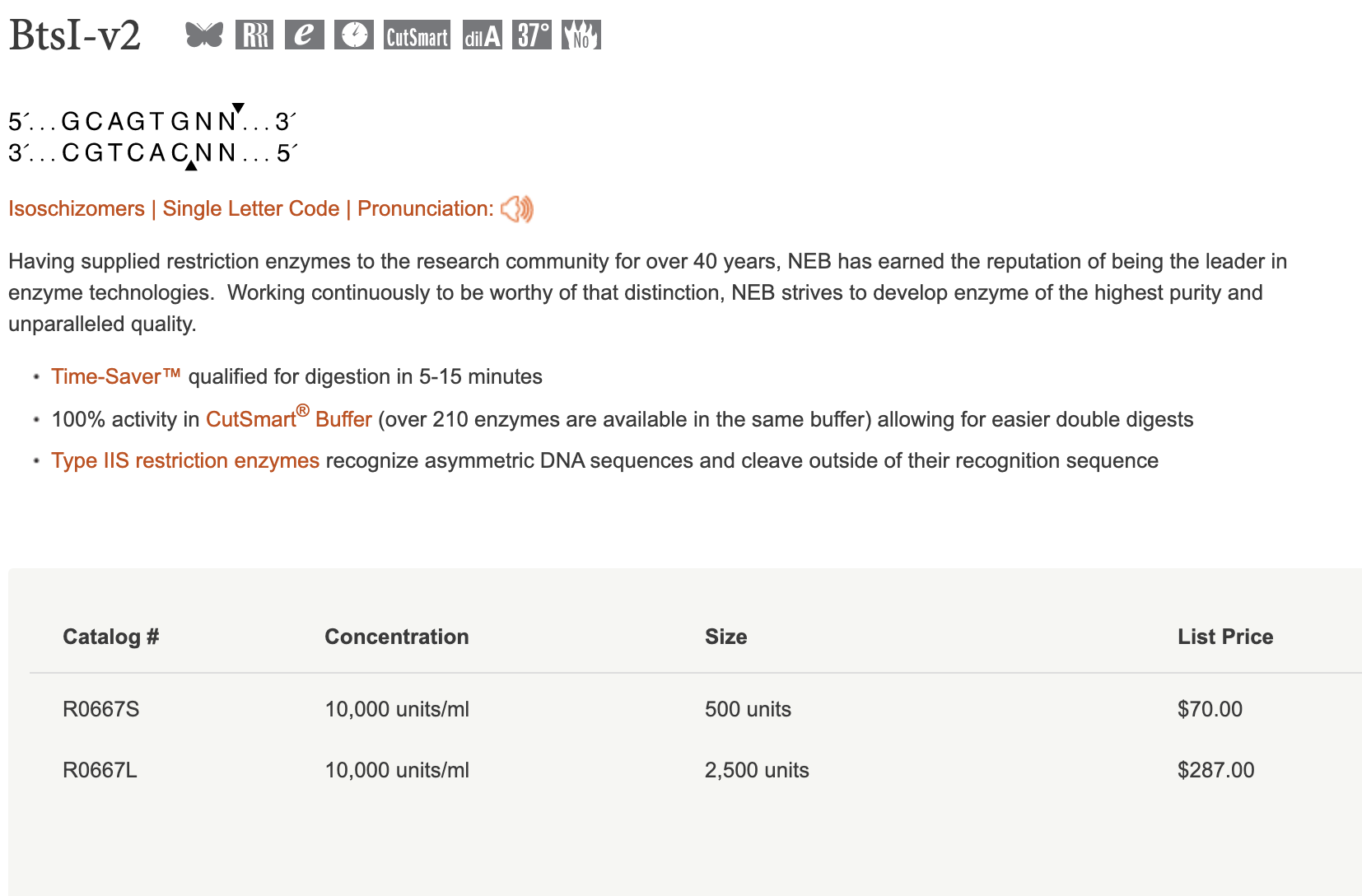
Further, both of these enzymes are quite cheap, and they both work in the Cutsmart buffer @ 37 degrees. I think that many different restriction enzymes could work for these two sites, for example nlaIII could work for the 5' site as well, but it wouldn't allow us to cleanly include a 'T' as a backup plan...but good to keep in mind for the future. Keep in mind that REs that work at temps > 50 degrees might cause the guide oligo do melt, leaving ssDNA, which would probably reduce the efficiency of the reaction.
For short recognition sequences, we can actually just find orthogonal kosuri primers that have the desired restriction site sequences. It would actually benefit this particular order to have longer sequences to be closer in length to the reg-seq constructs. The only issue is that when we purify / clean up this reaction we will be trying to purify a 128 bp oligo from 20 bp oligo, which already may be difficult. The longer those flanking oligos are the harder that step may be...that said these pcr handles should have no homology to genome and hopefully wouldn't affect anything even if they are electroporated directly into cell.
So, this notebook will find orthogonal primers that match our restriction sites (and add a 'CT' to BtsI-V2 site) to append to the ORBIT sequences of interest.